
Linux is readily establishing itself in the biomedical field as a powerful and reliable system for research computing.
The boom in biomedical research over the past decade has occurred as a result of increased demands for better health care and greater understanding of the biomolecular mechanisms responsible for many diseases. Fueling many of the scientific advances in medicine have been the corresponding developments in computational processing power and speed. In addition to the large-scale data collection and processing required to understand complex biological and physiological processes, our ability to analyze and model medical phenomena at microscopic and macroscopic levels is reliant upon the systems used for these computationally intensive jobs. Linux systems are receiving recognition as ideal workhorses for performing many of these tasks.
To better understand the emerging biomedical applications of Linux, it is first necessary to examine the typical computational requirements and resources of many biomedical research centers. The popular perception of biology and medicine is that they are wet and gushy sciences, involving little or no quantitative or numerical analysis. In fact, this couldn't be farther from the truth. There is a concerted effort among scientists to bring the biomedical sciences into the realm of other quantitative disciplines. This effort is illustrated by the recent emergence or increasing prominence of fields such as bioinformatics and genomic-sequence databases, biomolecular modeling and computer-aided drug design, diagnostic medical imaging, epidemiology and biostatistical analyses of patient populations, as well as medical informatics and patient record databases.
The systems used by researchers in these fields span a spectrum of different platforms. One typically finds an abundant assortment of Intel and Power PC-based systems running the Windows and Mac operating systems for performing basic tasks such as text processing, routine data analysis, e-mail and Internet access. In addition, the recent trend among manufacturers has been to interface a broad range of biomedical research equipment with PCs for the ease of GUI instrument control and data collection. The choice of which platform should be employed for such interfaces is usually the decision of the equipment manufacturer and not the researcher.
Laboratories may also contain a few Unix workstations which are used to process large jobs, drive complex instrumentation or function as servers. Typically these machines are dedicated to specific tasks and are unavailable for general use.
One unfortunate aspect of this heterogeneous situation is that it creates headaches for system administrators. Such a wide variety of systems performing a multitude of different tasks and operated by a large number of users with a wide range of computer abilities, frequently spells disaster.
Linux serves as an ideal system for biomedical research laboratories, where there is an abundance of PCs, strict limits on the allocation of research funds and a need for significant processing power in conjunction with straightforward and flexible system customization. The stability of Linux, its cost-effectiveness and large user community make it attractive to both novice and experienced system administrators confined by tight research computing budgets. In addition, the ability of the various incarnations of Linux to run on a wide variety of platforms provides a uniform OS and networking capabilities for the mixture of different machines already in existence at many biomedical research centers.
Utilization of Linux systems is emerging within the Biomolecular Structure Group here at Harvard Medical School. Intel-based systems running Red Hat Linux have recently been implemented for simultaneous control and data collection from x-ray diffraction instruments (Figure 1). Individual data sets obtained with these instruments are about 100MB in size and are used to understand the atomic structure of biomolecules and for drug-design studies at the molecular level. Although the source code for driving and collecting data on the x-ray instrument had to be recompiled for Linux and the kernel was rebuilt, the result was a very stable system for performing this specialized and complex task. In this instance, the Linux system has successfully replaced a more expensive Silicon Graphics workstation.

Figure 1. X-Ray Diffraction Equipment
Granted, the above application of Linux is rather specific, but Linux is also gaining more widespread usage among individual researchers. Installation of Linux on PCs in conjunction with either the Windows or Mac operating systems gives users the choice of the appropriate system with which to complete a job, while sitting at the same machine. For example, in a laboratory with an abundance of Macintoshes, the recently developed MkLinux for Power PC has proven to be beneficial in providing greater access to remote Unix systems and applications. More individuals are now able to use popular GUIs for genomic databases, bioinformatics applications, and biomolecular visualization software located on remote servers, while taking advantage of the multitasking capabilities of Linux (Figure 2). In addition, through implementation of a simple Tcl/Tk script, general users have the ability to reboot the machine back into Mac OS with a click of a button. In some cases, individuals are using the Linux systems as nothing more than X terminals. However, this use alone is valuable, since it provides increased access to applications located on a limited number of servers without having to purchase additional X terminals or X terminal emulation software.
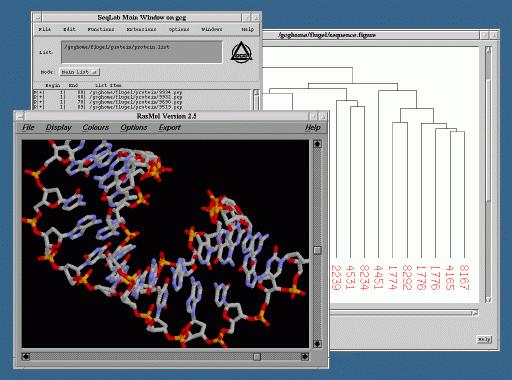
Figure 2. Multitasking with Linux
Linux is bound to have a more prominent role in biomedical research computing in the future. The recent installations of Linux systems described above have already been recognized as highly successful and even superior to other systems, in terms of both price and performance. As the computing demands of biomedical research increase, scientists, system administrators, software developers and instrument manufacturers will all be considering Linux.